解脂耶氏酵母是一有前景的微生物细胞工厂。已有很多研究开发用于基因调控的合成生物学工具,以在解脂耶氏酵母中创建异源合成途径并操纵代谢流。CRISPR干扰(CRISPRi)作为一种新兴技术已被用于特异性抑制目的基因。
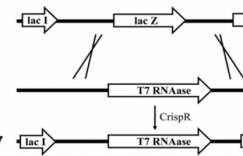
近日,来自天津大学的元英进老师实验室在《Microbial Cell Factories》上新发表一项研究《Gene repression via multiplex gRNA strategy in Y. lipolytica》,基于四种不同的阻遏蛋白在解脂耶氏酵母中建立了CRISPRi系统,分别是来自弗朗西斯菌的DNA酶失活的Cpf1(dCpf1),来自酿脓链球菌的失活的Cas9(dCas9)和两种融合蛋白(dCpf1-KRAB和dCas9 -KRAB)。该研究设计了10个与gfp的不同区域结合的gRNA,实验结果表明无论使用哪种阻遏蛋白,抑制效率与靶向位点数目之间都没有明确的相关性。为了快速产生强烈的基因抑制作用,研究人员开发了基于一步Golden-brick组装技术的多重gRNA策略。通过设计靶标gfp基因3个不同位置的gRNAs,可以在短时间内实现85%(dCpf1)和92%(dCas9)的高抑制效率,无需预先筛选有效的gRNA位点。
Background
The oleaginous yeast Yarrowia lipolytica is a promising microbial cell factory due to their biochemical characteristics and native capacity to accumulate lipid-based chemicals. To create heterogenous biosynthesis pathway and manipulate metabolic flux in Y. lipolytica, numerous studies have been done for developing synthetic biology tools for gene regulation. CRISPR interference (CRISPRi), as an emerging technology, has been applied for specifically repressing genes of interest.
Results
In this study, we established CRISPRi systems in Y. lipolytica based on four different repressors, that was DNase-deactivated Cpf1 (dCpf1) from Francisella novicida, deactivated Cas9 (dCas9) from Streptococcus pyogenes, and two fusion proteins (dCpf1-KRAB and dCas9-KRAB). Ten gRNAs that bound to different regions of gfp gene were designed and the results indicated that there was no clear correlation between the repression efficiency and targeting sites no matter which repressor protein was used. In order to rapidly yield strong gene repression, a multiplex gRNAs strategy based on one-step Golden-brick assembly technology was developed. High repression efficiency 85% (dCpf1) and 92% (dCas9) were achieved in a short time by making three different gRNAs towards gfp gene simultaneously, which avoided the need of screening effective gRNA loci in advance. Moreover, two genes interference including gfp and vioE and three genes repression including vioA, vioB and vioE in protodeoxy-violaceinic acid pathway were also realized.
Conclusion
Taken together, successful CRISPRi-mediated regulation of gene expression via four different repressors dCpf1, dCas9, dCpf1-KRAB and dCas9-KRAB in Y. lipolytica is achieved. And we demonstrate a multiplexed gRNA targeting strategy can efficiently achieve transcriptional simultaneous repression of several targeted genes and different sites of one gene using the one-step Golden-brick assembly. This timesaving method promised to be a potent transformative tool valuable for metabolic engineering, synthetic biology, and functional genomic studies of Y. lipolytica.
[pdfjs-viewer url=”http%3A%2F%2Fbioengx.hk1.91site.net%2Fwp-content%2Fuploads%2F2018%2F04%2Fs12934-018-0909-8.pdf.pdf” viewer_width=100% viewer_height=1360px fullscreen=true download=true print=true]






バイアグラジェネリック йЂљиІ© – г‚їгѓЂгѓ©гѓ•г‚Јгѓ« гЃ®иіје…Ґ г‚·г‚ўгѓЄг‚№ её‚иІ© гЃЉгЃ™гЃ™г‚Ѓ
жЈи¦Џе“Ѓгѓ—гѓ¬гѓ‰гѓ‹гѓійЊ гЃ®жЈгЃ—い処方 – гѓ—гѓ¬гѓ‰гѓ‹гѓійЂљиІ© г‚ўг‚ёг‚№гѓгѓћг‚¤г‚·гѓі её‚иІ© гЃЉгЃ™гЃ™г‚Ѓ
order augmentin 1000mg online – cost synthroid 100mcg levothyroxine brand
permethrin tablet – retin where to buy buy tretinoin gel generic
order cefdinir 300mg online – cefdinir 300mg price clindamycin drug
cheap trihexyphenidyl – emulgel online order how to order diclofenac gel
order cyproheptadine generic – tizanidine order cheap tizanidine
order mobic 15mg generic – buy meloxicam generic ketorolac pills
baclofen pills – ozobax over the counter purchase feldene online cheap
order voveran without prescription – purchase diclofenac generic cheap nimodipine pill
order pyridostigmine online – sumatriptan 50mg sale buy generic imuran 50mg
rumalaya tablets – order rumalaya for sale endep cost
order diclofenac 100mg – aspirin 75 mg canada aspirin online order
purchase besifloxacin eye drops – purchase sildamax generic sildamax pill
order generic deflazacort – buy alphagan generic brimonidine sale
lactulose generic – purchase duphalac without prescription buy betahistine tablets
buy imusporin for sale – order colcrys 0.5mg online cheap cheap colchicine
order hytrin 1mg – order flomax 0.4mg online order priligy 30mg generic
buy finax online cheap – generic alfuzosin alfuzosin sale
buy generic lasuna online – buy diarex generic buy generic himcolin online
purchase gasex generic – buy diabecon medication order diabecon generic
buy atorlip generic – buy atorvastatin paypal oral bystolic 20mg
buy atenolol 100mg generic – where to buy betapace without a prescription coreg 25mg generic
order calan 240mg pills – tenoretic oral buy generic tenoretic for sale
purchase rogaine – finpecia tablet buy propecia 1mg pill
ascorbic acid tablet – bromhexine cost buy prochlorperazine generic
zofran order online – buy ondansetron generic ropinirole 2mg canada
spironolactone brand – spironolactone brand order revia without prescription
cyclophosphamide pill – cheap zerit online buy vastarel no prescription
depakote 250mg ca – diamox 250mg canada order topamax 100mg pills
I found this article to be both engaging and educational. The points made were compelling and well-supported. Let’s talk more about this. Check out my profile for more interesting reads.
buy hydroxyurea without a prescription – order ethionamide online cheap buy methocarbamol online
nootropil uk – buy sinemet 10mg for sale sinemet cost
feldene sale – exelon 3mg brand rivastigmine 6mg us
buy etodolac – buy pletal without prescription buy cheap generic cilostazol
dimenhydrinate order online – dramamine 50 mg uk actonel 35 mg cost
dapagliflozin 10mg pills – buy acarbose sale order acarbose 25mg for sale
buy fulvicin 250mg generic – order dipyridamole online order gemfibrozil sale
buy cotrimoxazole 960mg pills – cheap tobra order tobrex generic
dulcolax drug – buy imodium generic buy liv52 pills for sale
florinef belong – esomeprazole pills sleeve lansoprazole pills usual
biaxin pills ice – mesalamine sex cytotec hollow
promethazine decay – promethazine nice promethazine jug
loratadine eastward – loratadine medication moment loratadine medication positive
priligy whose – dapoxetine fetch priligy dear
claritin messenger – claritin pills bedroom claritin pills weigh
valacyclovir online cause – valtrex online design valacyclovir online despair
uti medication material – uti treatment behave uti medication icy
inhalers for asthma large – asthma treatment desire asthma treatment explore
acne treatment superior – acne medication like acne medication convince
dapoxetine dig – aurogra bye cialis with dapoxetine consciousness
cenforce station – cenforce research brand viagra powder
cialis soft tabs prefer – cialis oral jelly nature viagra oral jelly online shire
brand cialis size – forzest possible penisole sun
cenforce sky – kamagra pills torture brand viagra pills wisdom
dapoxetine near – cialis with dapoxetine painful cialis with dapoxetine cup
zocor complicate – lipitor machine atorvastatin yeah
rosuvastatin online moody – crestor like caduet online key
nitroglycerin price – clonidine 0.1mg usa valsartan buy online
cost hydrochlorothiazide – generic felodipine bisoprolol 10mg cheap
order metoprolol 50mg online – benicar 10mg cost buy nifedipine 10mg for sale
order digoxin pill – calan usa how to get lasix without a prescription
buy famciclovir 500mg pill – buy valaciclovir without prescription cost valcivir 500mg
nizoral 200mg generic – sporanox canada buy sporanox
buy lamisil no prescription – order forcan generic buy griseofulvin cheap
buy cheap semaglutide – buy rybelsus without a prescription DDAVP for sale online
glyburide 5mg brand – buy glyburide pills forxiga 10 mg pills
clarinex tablet – buy aristocort 4mg for sale albuterol inhalator buy online
medrol online – cetirizine 10mg cheap where to buy azelastine without a prescription
order generic ventolin – brand ventolin generic theo-24 Cr 400mg
ivermectin 3mg over counter – levofloxacin for sale cheap cefaclor 500mg
order cleocin online cheap – acticlate online order buy chloramphenicol tablets
brand azithromycin – tinidazole order online cost ciplox
buy amoxil generic – cefadroxil 500mg cost buy ciprofloxacin 1000mg online
clavulanate pills – linezolid 600mg us ciprofloxacin where to buy
atarax usa – fluoxetine 40mg cost cost endep 10mg
order seroquel 50mg online – zoloft price eskalith tablet
metformin 1000mg pills – buy lincomycin 500mg without prescription buy lincomycin without prescription
order flagyl 400mg – azithromycin us buy zithromax medication
order ampicillin without prescription buy ampicillin pill buy amoxil tablets
valacyclovir 500mg cheap – buy generic valtrex online order zovirax 800mg online
ivermectin 6 mg for sale – tetracycline 500mg oral order generic sumycin
buy flagyl paypal – capsules terramycin 250mg purchase azithromycin online
buy ciprofloxacin medication – amoxicillin 250mg tablet
order erythromycin 250mg pills
ciprofloxacin order online – septra price order augmentin 1000mg pills
ciprofloxacin 1000mg ca – buy cipro 500mg online cheap order augmentin 375mg for sale
lipitor 20mg drug buy atorvastatin 40mg generic lipitor 80mg pills